20/21 deaths in a Chinese CRISPR PD-1 k/o trial. Our cells just cant take so much genomic damage.
As I show in this nanopore pre-print, there are off-targets with 7 mismatches. And in this BCL11a stem cell k/o, off-targets with 8 mismatches.
And with 8 mismatches (maybe more) allowed, there are 100k possible off-targets for any gRNA in most any genome. It wont always cut, but even with low efficiency the final expected numbers will be significant.
And this seminal 2014 paper showing “mismatches at dCas9 binding sites can be as high as 10” tells you why.
Plus, we dont really have a good tech to find it – as I show in this paper biorxiv rejected on TALEN edited cows.
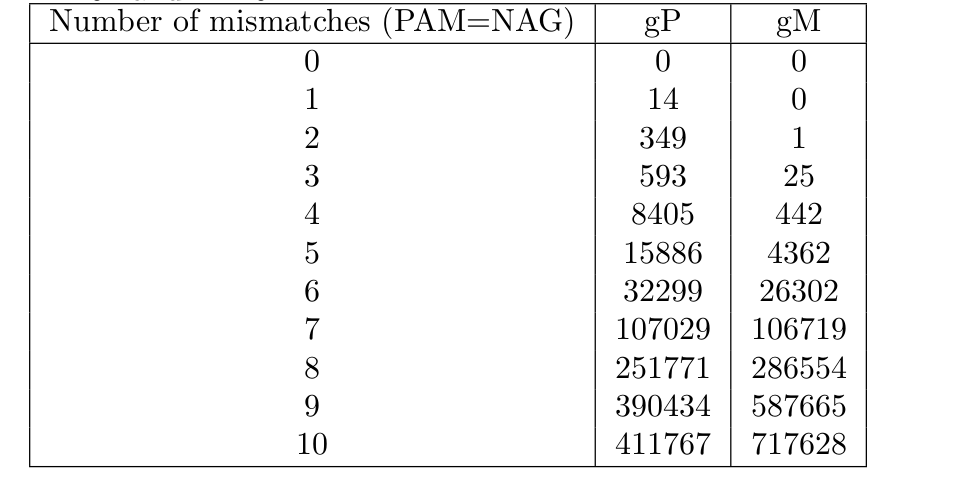
Just one PAM – and that too with the constraint of less than 2 mismatches in the 12 PAM-proximal (which is a true constraint).
Once a large number of DSBs are made p53 would be activated – killing cells, and thus selecting p53 deficient cells, which would be subject to genotoxic damage.
Recent studies from other groups showing showing significant off-targets
- Frequent loss-of-heterozygosity in CRISPR-Cas9-edited early human embryos
- “Allele-Specific Chromosome Removal after Cas9 Cleavage in Human Embryos”
- Stem cells
- Pacbio
- Cosmo – the calf : “Cosmo’s SRY pile-up is precisely the kind of unforeseen consequence that Crispr critics worry about when it comes to the risks of gene editing. “